Appearance
Main Menus
The Main Menu is the central interface for managing structures, ligands, workspaces, collaboration, and settings in Nanome v2. It is accessed via the Wrist Menu and is personalized to each user in the session. The Main Menu contains five tabs across the top: Components, Ligands, Workspaces, Collaboration, and Settings. Additional panels — Scenes, Sequence, and Tools — are accessible from the edges of the interface.
Components Tab
The Components Tab is where you manage all loaded structures and their visual representations.
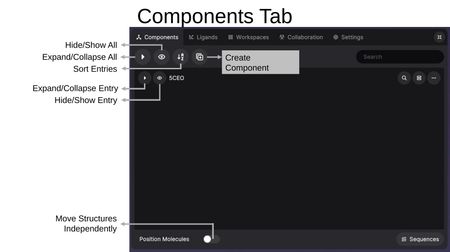
Toolbar
The top toolbar provides global controls:
- Expand/Collapse All: Toggle all entries open or closed.
- Hide/Show All: Toggle visibility of all structures at once.
- Sort Entries: Reorder entries alphabetically.
- Create Component: Create a new custom component from a selection.
- Search: Filter entries by keyword.
- Expand/Contract Menu: Resize the menu panel using the icon in the top-right corner.
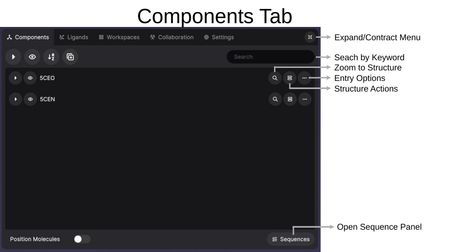
Each entry row has its own set of controls:
- Expand/Collapse Entry: Show or hide the components within an entry.
- Hide/Show Entry: Toggle the visibility of the entry in the 3D view.
- Zoom to Structure (magnifying glass): Center the view on this structure.
- Entry Options (clipboard icon): Access structure actions menu.
- More Options (...): Rename or delete the entire entry.
At the bottom of the panel, Position Molecules allows you to move structures independently when toggled on, and Sequences opens the Sequence Panel.
Entries and Components
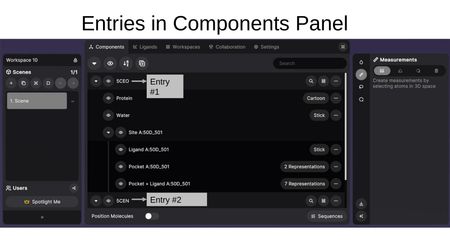
Structures are organized as entries (e.g., PDB codes like 5CEO, 5CEN). Each entry contains components, which are sub-selections of the structure with their own representation settings.
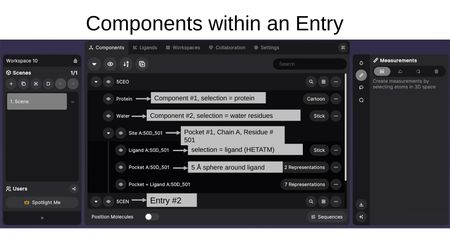
For example, an entry might contain:
- Protein — displayed as Cartoon
- Water — displayed as Stick
- Site A:50D_501 — a binding site group containing:
- Ligand A:50D_501 — the ligand, shown as Stick
- Pocket A:50D_501 — the 5 Å sphere around the ligand
- Pocket + Ligand A:50D_501 — combined view with multiple representations
Representations and Color Options
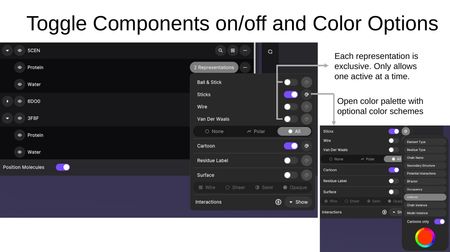
Clicking the representation button on a component opens the representation panel. Each representation type is exclusive — only one atomistic representation can be active at a time per component. The color palette icon opens color scheme options.
Atomistic Representations:
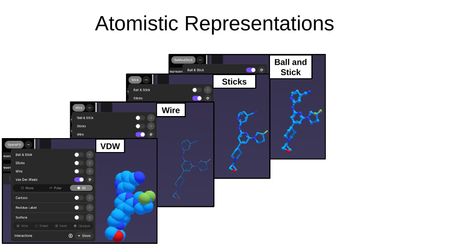
- Ball and Stick — atoms as spheres connected by sticks
- Sticks — bonds shown as sticks with no atom spheres
- Wire — thin lines representing bonds
- VDW (Van der Waals) — space-filling model showing atomic radii
Additional options include hydrogen display modes: None, Polar, or All.
Macromolecular Representations:
- Cartoon — secondary structure ribbons (helices, sheets, loops)
- Residue Label — labels for each residue
- Surface — molecular surface with four density levels:

- Wire — wireframe mesh surface
- Sheer — transparent surface
- Semi — semi-transparent surface
- Opaque — solid surface
Interaction Types:
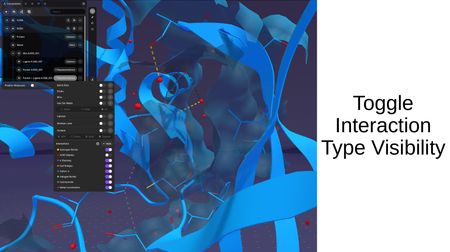
When interactions are computed between structures, you can toggle the visibility of individual interaction types: Hydrogen Bonds, VDW Clashes, π Stacking, Salt Bridges, Cation-π, Halogen Bonds, Hydrophobic, and Metal Coordination.
Color Schemes:
The color palette provides schemes including Element Type, Residue Type, Chain Name, Secondary Structure, Potential Interactions, BFactor, Occupancy, Uniform, Chain Instance, and Model Instance. A "Carbons only" toggle applies the color scheme to carbon atoms only while preserving element colors for heteroatoms.
Structure Actions

Clicking the Entry Options button on an entry opens the Structure Actions menu with the following options:
- Align proteins to this — Align other structures to this entry. Select target entries and specific chains (All, A, B, etc.), then click Align. After alignment, RMSD values are displayed between entries.
- Create component — Create a custom component by specifying residue ranges, types, amino acids, or nucleotides.
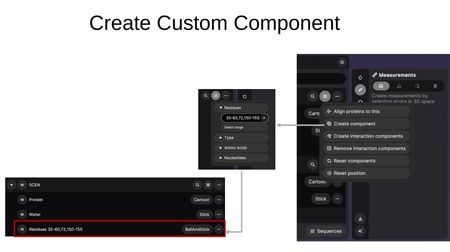
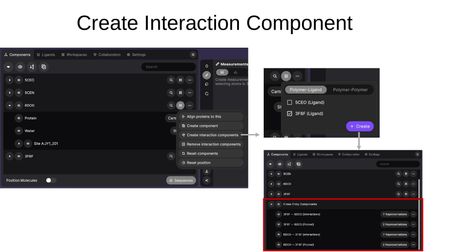
- Create interaction components — Compute interactions between structures. Choose between Polymer-Ligand and Polymer-Polymer modes, select the target entry, and click Create. This generates Cross-Entry Components showing interactions and pocket regions between the selected structures.
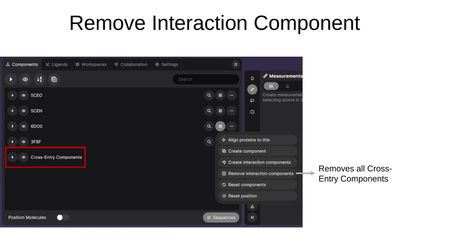
- Remove interaction components — Removes all Cross-Entry Components from the workspace.
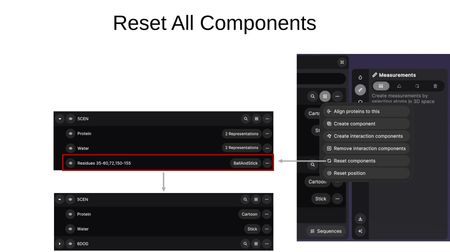
- Reset components — Removes all custom components and restores entries to their default component breakdown (e.g., Protein + Water).
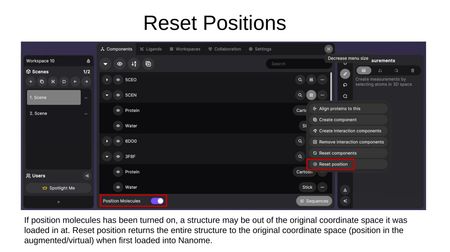
- Reset position — Returns a structure to its original coordinate space. This is useful if Position Molecules was toggled on and a structure was moved out of its original position.
Entry More Options
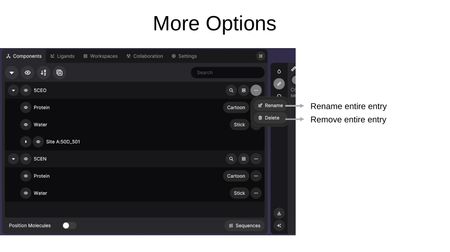
Clicking the (...) button on an entry provides:
- Rename — Change the display name of the entry.
- Delete — Remove the entire entry from the workspace.
Ligands Tab
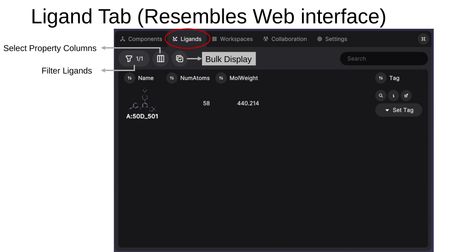
The Ligands Tab provides a table view for viewing, rating, and managing ligands across all loaded structures. The interface resembles the MARA web interface.
Toolbar
- Filter Ligands — Filter the ligand list by criteria.
- Select Property Columns — Choose which properties to display as columns (RDKit-calculated properties and SDF metadata from source files).
- Bulk Display — Simultaneously display multiple models from a multi-model file.
- Search — Search for a ligand by name.
Ligand Table
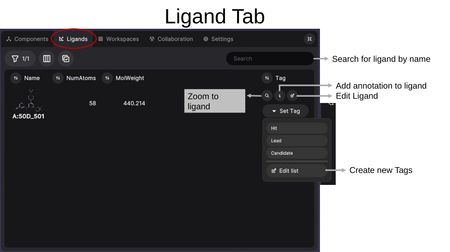
Each ligand row displays the 2D structure preview along with selected property columns (e.g., Name, NumAtoms, MolWeight). Per-ligand actions include:
- Zoom to ligand (magnifying glass) — Center the 3D view on this ligand.
- Add annotation (i) — Add notes or annotations to the ligand.
- Edit Ligand (pencil) — Open the ligand in the Builder Tool for editing.
- Set Tag — Assign tags such as Hit, Lead, or Candidate. You can also create custom tags via "Edit list".
Property Columns
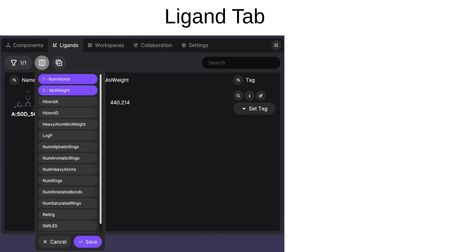
Available properties include RDKit-calculated values (NumAtoms, MolWeight, HbondA, HbondD, HeavyAtomMolWeight, LogP, NumAliphaticRings, NumAromaticRings, NumHeavyAtoms, NumRings, NumRotatableBonds, NumSaturatedRings, Rating, and SMILES) as well as any SDF metadata from the source file.
Workspaces Tab
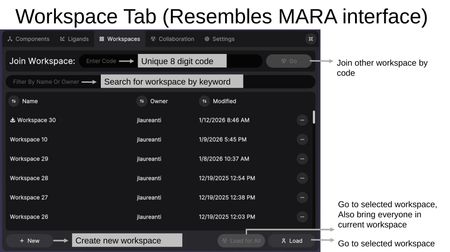
The Workspaces Tab allows you to manage, create, and navigate between workspaces. The interface resembles the MARA web interface.
Features
- Join Workspace — Enter a unique 8-digit workspace code to join another user's workspace.
- Search — Filter workspaces by name or owner.
- Workspace List — Shows all accessible workspaces with Name, Owner, and Modified date. Each workspace has an options menu (...).
- + New — Create a new workspace.
- Load for All — Go to the selected workspace and bring everyone in the current session with you. Users will be prompted to assign permissions if they don't already have them.
- Load — Go to the selected workspace for yourself only, leaving other participants in their current workspace.
Collaboration Tab
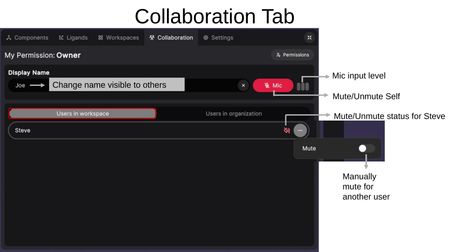
The Collaboration Tab manages users, audio, and permissions within a workspace.
User Settings
- Display Name — Change the name visible to other users in the session.
- Mic — Toggle your microphone on/off. The mic input level indicator shows when audio is being transmitted.
Users in Workspace
Lists all users currently in the workspace. For each user you can:
- View their mute/unmute status.
- Click (...) to manually mute another user.
Users in Organization
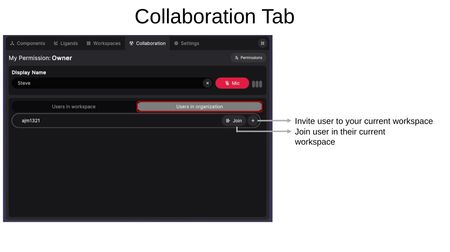
Lists all users in your organization who are currently online. For each user you can:
- Join — Join that user in their current workspace.
- (+) — Invite that user to your current workspace.
Permissions
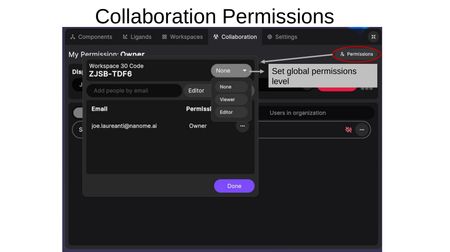
Click the Permissions button (top-right) to open the permissions dialog. The dialog shows the Workspace Code and provides:
- Global permissions level — Set the workspace to None, Viewer, or Editor for anyone with the code.
- Add people by email — Invite specific users by email and set their permission level (Editor or Viewer). This must be an exact email match.
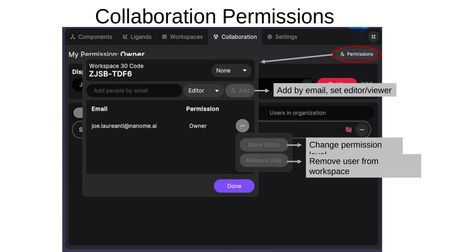
For individual users already in the workspace, click (...) to:
- Change permission level — Switch between Editor and Viewer.
- Remove user — Remove the user from the workspace.
Settings Tab
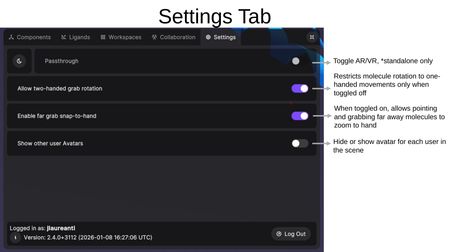
The Settings Tab provides environment and interaction preferences.
- Passthrough — Toggle between AR (mixed reality passthrough) and VR modes. Available on standalone headsets only.
- Allow two-handed grab rotation — When toggled off, restricts molecule rotation to one-handed movements only.
- Enable far grab snap-to-hand — When toggled on, allows pointing at and grabbing far-away molecules to bring them closer.
- Show other user Avatars — Hide or show avatar representations for each user in the scene.
The bottom of the Settings Tab displays the currently logged-in username, the Nanome version number, and a Log Out button.
Scenes Panel
The Scenes Panel is accessible from the left side of the interface and manages scene views within a workspace.
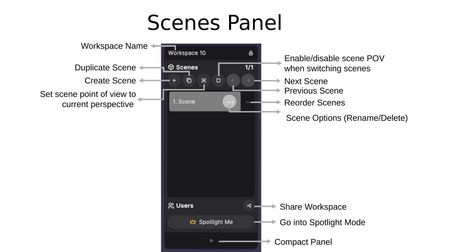
Controls
- Workspace Name — Displayed at the top of the panel, along with a lock icon indicating permission status.
- Create Scene (+) — Add a new scene to the workspace.
- Duplicate Scene — Copy the current scene with all its representations and settings.
- Set Scene POV — Save your current perspective as the point of view for this scene.
- Enable/Disable Scene POV — Toggle whether switching scenes automatically adjusts your viewpoint.
- Previous / Next Scene — Navigate between scenes using the arrow buttons.
- Scene Options (...) — Access rename and delete options for individual scenes.
- Reorder Scenes (=) — Drag to rearrange scenes in the list.
Users Section
At the bottom of the Scenes Panel, the Users section shows who is currently in the workspace. Each user is assigned a personal color, displayed on their name tag and their crown icon. A Share Workspace button allows you to invite others, and a Compact Panel toggle (>>) collapses the panel.
Multi-User Scene Indicators
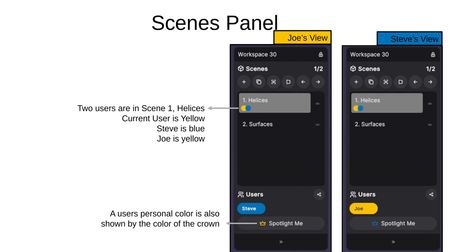
When multiple users are in a workspace, colored dots on each scene indicate which users are currently viewing that scene. Each user's personal color corresponds to their name tag in the Users section.
Spotlight Mode
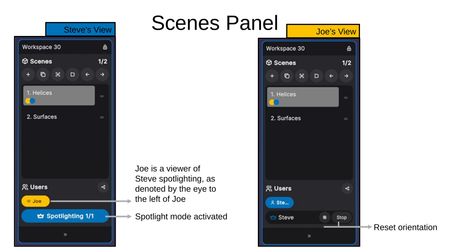
Clicking Spotlight Me activates Spotlight Mode, making your view the shared perspective for all followers.
- Spotlighter's View — The "Spotlighting 1/1" indicator shows Spotlight Mode is active and how many users are following. Followers are listed with an eye icon next to their name.
- Follower's View — Followers see the spotlighter's name with a crown icon, along with a Reset Orientation button and a Stop button to exit follow mode.
Sequence Panel
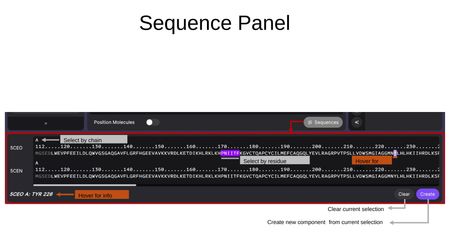
The Sequence Panel is opened by clicking the Sequences button at the bottom-right of the Components Tab. It displays the amino acid sequences of all loaded structures in a horizontal, scrollable view.
Features
- Select by chain — Click the chain letter (e.g., "A") to select an entire chain.
- Select by residue — Click individual residues in the sequence to select them. Selected residues are highlighted.
- Hover for info — Hovering over a residue shows its details (e.g., "5CEO A: TYR 226") at the bottom of the panel.
- Clear — Clear the current selection.
- Create — Create a new component from the current residue selection.
When a component is active in the sequence panel, an Update button appears to update the existing component with the current selection.
Tools Panel
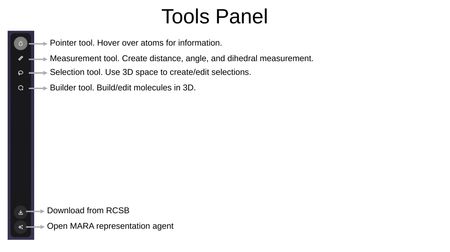
The Tools Panel is a vertical toolbar on the right side of the interface providing access to interaction tools. From top to bottom:
- Pointer Tool — Hover over atoms to display information.
- Measurement Tool — Create distance, angle, and dihedral measurements.
- Selection Tool — Use 3D space to create and edit selections.
- Builder Tool — Build and edit molecules in 3D.
- Download from RCSB — Import structures directly from the RCSB Protein Data Bank.
- Open MARA — Launch the MARA representation agent.
Selection Tool
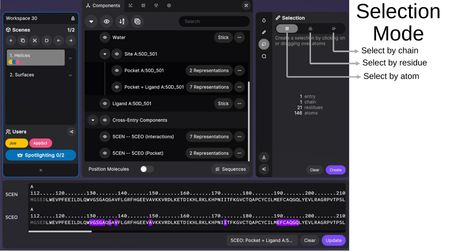
The Selection Tool allows you to create selections by clicking on or dragging over atoms in 3D space. Selection granularity options include:
- Select by chain — Select entire chains.
- Select by residue — Select individual residues.
- Select by atom — Select individual atoms.
The Selection Panel shows information about the current selection (entry count, chains, residues, atoms) and provides:
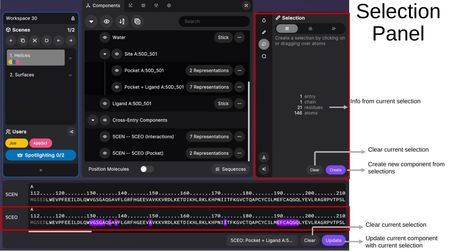
- Clear — Clear the current selection.
- Create — Create a new component from the selection.
In the Sequence Panel, when a component is selected, you can also Update the component with the current selection or Clear it.
Measurement Tool
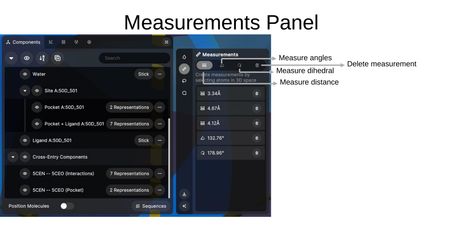
The Measurement Tool creates measurements by selecting atoms in 3D space. Measurement types include:
- Distance — Measure the distance between two atoms (displayed in Ångströms).
- Angle — Measure the angle between three atoms (displayed in degrees).
- Dihedral — Measure the dihedral angle between four atoms (displayed in degrees).
Each measurement appears as a row in the Measurements Panel with a delete button to remove individual measurements.
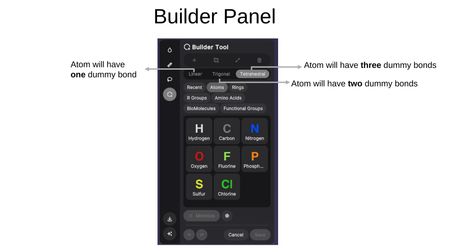
Builder Tool

The Builder Tool enables 3D molecular construction and editing. Upon activation, you can choose to:
- Create New — Start building a new molecule from scratch.
- Edit existing — Select an existing ligand to modify.
The Builder Panel provides the following modes:
- Add atom (+) — Place individual atoms. Geometry options determine the number of dummy bonds: Linear (one), Trigonal (two), or Tetrahedral (three).
- Add by group — Insert pre-built groups from categories including: Recent, Atoms, Rings, R Groups, Amino Acids, BioMolecules, and Functional Groups.
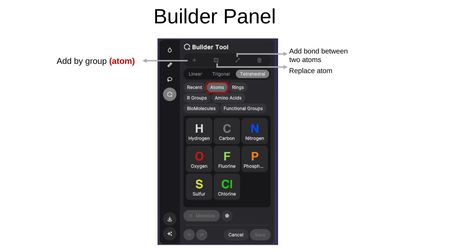
- Add bond — Create a bond between two existing atoms.
- Replace atom — Swap one atom type for another.
- Delete atom — Remove atoms from the structure.
Available Atoms: Hydrogen (H), Carbon (C), Nitrogen (N), Oxygen (O), Fluorine (F), Phosphorus (P), Sulfur (S), and Chlorine (Cl).
Minimization
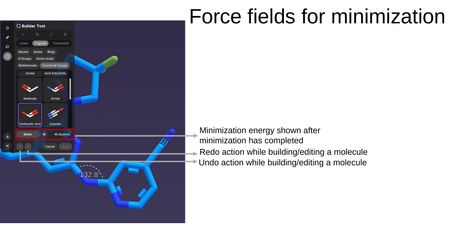
While in the Builder Tool, clicking Minimize runs a force field minimization on the structure. After minimization completes:
- The minimization energy is displayed (e.g., 46.6 kJ/mol).
- Undo/Redo buttons allow you to step through edits made during the building session.
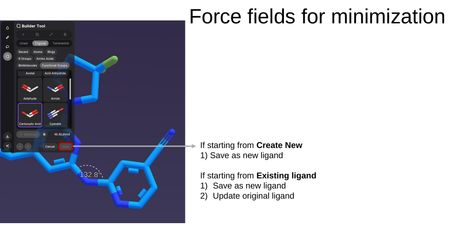
When saving, the options depend on your starting point:
- Starting from Create New: Save as new ligand.
- Starting from an existing ligand: Save as new ligand, or Update original ligand.