Appearance
Antibodies Representation
The Antibody Representation plugin adds the IMGT color scheme, labels antibody regions on a selected protein, and highlights CDR loops. A table of Antibody Regions allows independent selection of CDR loops for calculations or representational changes. Importantly, only antibodies and fragments of antibodies are modified; the representation of antigens in the complex is unchanged.
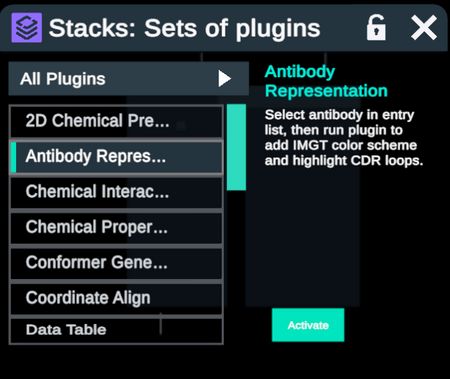 | The plugin is accessible through the stacks menu. Select the antibody structure in the Entry list, click Activate and then Run. The structure will update with heavy and light chains colored in grays and CDR loops colored according to the IMGT scheme. |
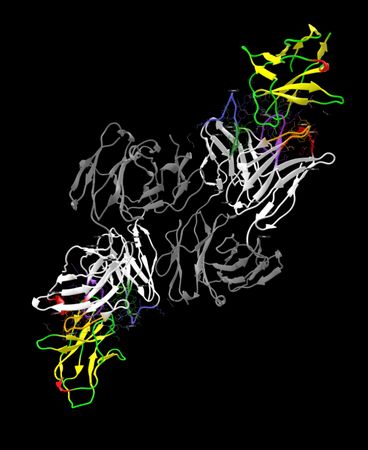 | 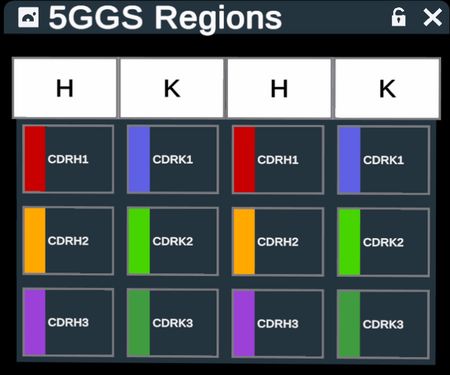 |
Output of run on PDB code 5GGS, PD-1 + pembrolizumab Fab and Regions Table. Note that only antibody portions are recolored and labeled; antigen protein remains as loaded
Select CDR loops from the table by clicking on the labeled buttons. The 3D structure will show selection highlights on the loops.
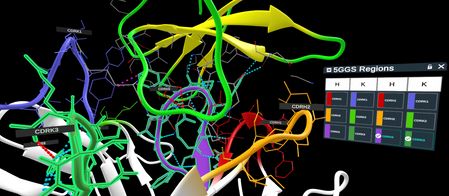
5GGS close up of the interface of PD-1 with the CDR loops of the antibody fragment, with 2 loops selected in the table and on the structure
Application of CDR Selections
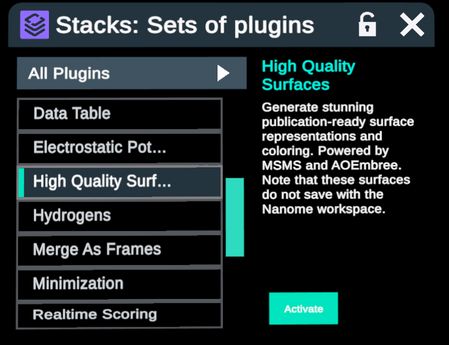
Selection of CDR loops from the Regions table can be used in conjunction with other Nanome menus and plugins. Some examples of plugins are Chemical Interactions and High Quality Surfaces.
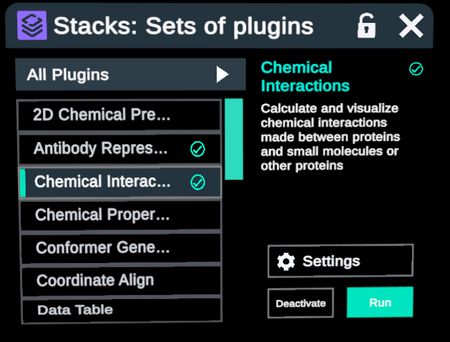 |  |
Calculate Chemical Interactions between the residues of selected CDR loops with proteins within 5 angstroms in 4 steps.
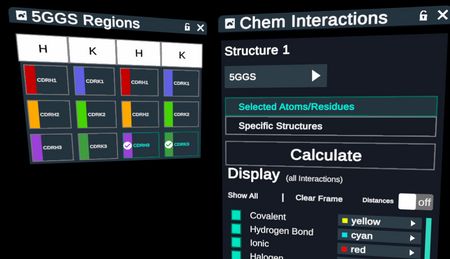
- Make selections in the Regions table
- Activate and Run the Chemical Interactions plugin from the stacks menu to launch the plugin menu
- In the menu, confirm the selection of your protein, the selection of “Selected Atoms/Residues”, and make any adjustment to what interactions to show.
- Click “Calculate”

Chemical Interactions made by the residues of 2 CDR loops
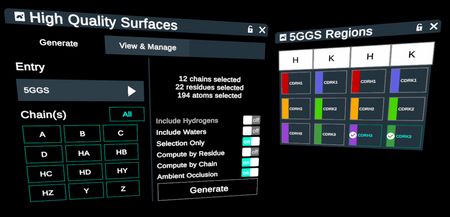
Generate High Quality Surfaces on any selection of CDR loops in 5 steps.
- Make selections in the Regions table
- Activate the High Quality Surfaces plugin from the stacks menu to launch the plugin menu
- In the menu, select the protein, and a chain that contains the selection or the entire protein.
- In the right panel of the menu, set the “selection only” toggle to “on”. This will apply surfaces to the selected CDR loops in the Regions table, as well as any other selections made on the structure using the select tool, entry list, or other menus.
- Click “Generate” and modify surface colors and opacity to purpose.
Citations
David Příhoda. (2021). AbNumber: Convenience Python APIs for antibody numbering and alignment using ANARCI (Version 0.3.2). Retrieved from https://github.com/prihoda/AbNumber/
Dunbar, J., & Deane, C. M. (2016). ANARCI: antigen receptor numbering and receptor classification. Bioinformatics, 32(2), 298-300. https://doi.org/10.1093/bioinformatics/btv552