Appearance
Main Menus
Entry List
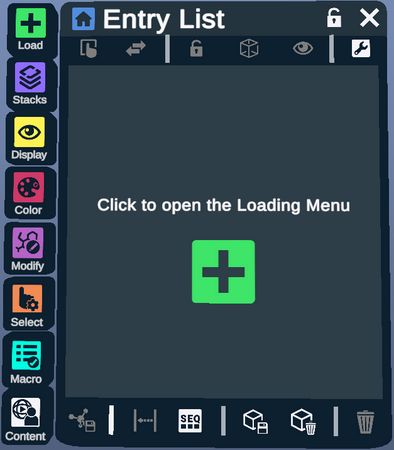
The Entry List is a list of all of the structures that have been loaded into the environment.
The Load, Plugin, Display, Color, Modify, Select, Macro, and Spatial Tutorial Menus can be displayed by clicking their respective icons located at the bottom of the Entry List.
Device Limiter
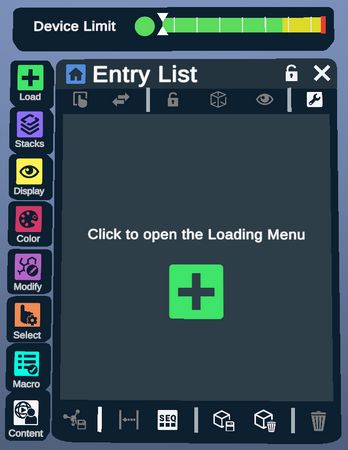
The Device Limiter is an indicator that shows how close the current amount of atoms is to hitting the limit of a standalone device's computing capability. This indicator only shows up when using a device in standalone mode (if applicable). When the amount of atoms approaches or hits the limit, the following prompts will show up.


Hierarchy Menu
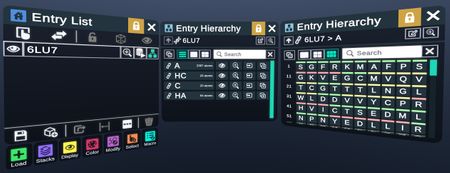
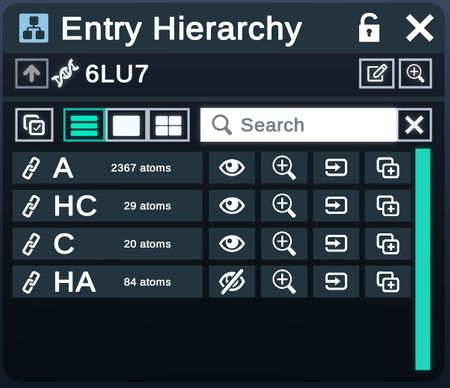
The Hierarchy Menu lists out the components of an entity, such as chains and residues, in hierarchical levels.
Different Browsing Modes (Small, Medium, Large)
Different Viewing Modes can be toggled to modify the substructure icon sizes in each menu.
Frame Data
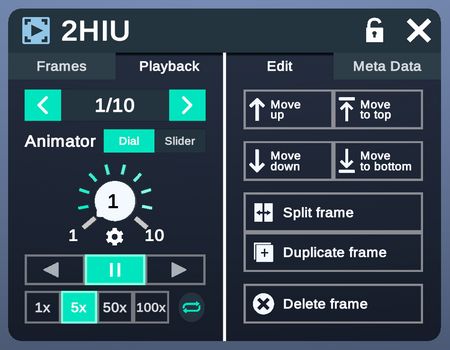
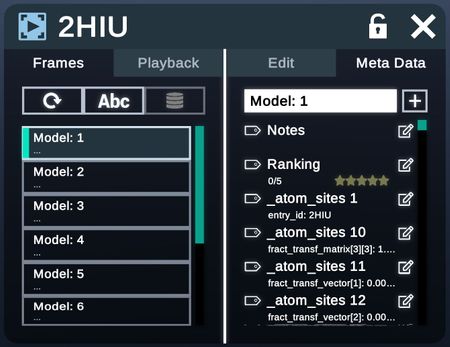
Frame data allows users to navigate and modify different frames of the same structure (Academic and Enterprise only).
Electron Density Maps

▶
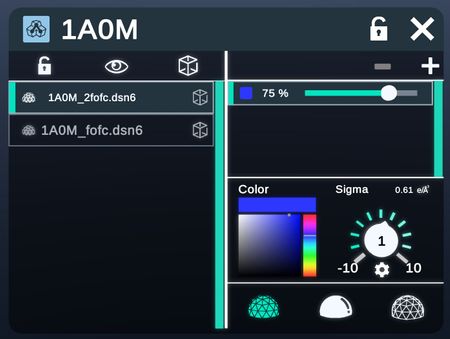
The Electron Density tab shows the configuration of viewing the Electron Density Map of the structure (Academic and Enterprise only).
Alternative Conformations Menu
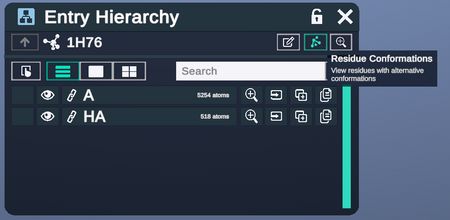
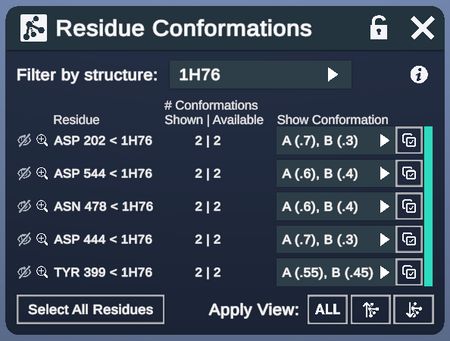
This menu specifies residue conformations.
Menu Actions
Action buttons on the Entry List provide quick access to common actions on the workspace.
| Action | Description | Additional Menu |
|---|---|---|
  | Select or deselect all present structures. | |
 | Invert current selection. | |
 | Lock or unlock selected entities. | |
 | Show or hide the box frame of selected entities. | |
 | Show or hide the selected entities. | |
 | Save the current workspace. | |
 | Discard the current workspace and create a new one. | |
 | Export the selected structures in a desired format. | 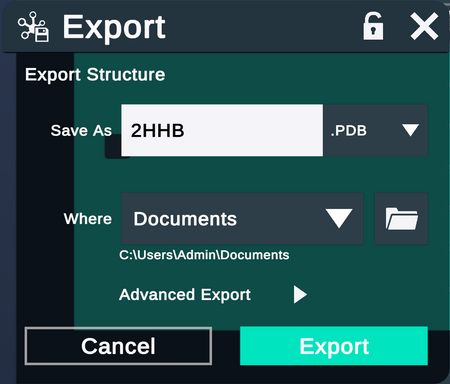 |
 | Align the selected entities and place them in the same position under the same coordinate system. Caution: this does not necessarily align the entities by their atom position, and the entities could end up not overlapping after the alignment. | |
 | View the sequences of multiple chains and align them. | 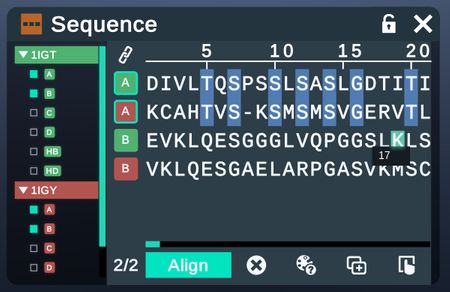 |
 | Delete the selected entities. |
Spatial ("Quick") Anchor
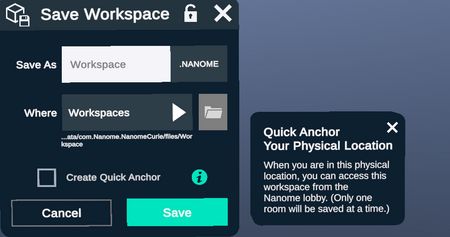
When saving a workspace, you have the option to create a spatial anchor for your current physical location. When you re-launch Nanome in this same physical location, you'll be able to quickly access this workspace from the Nanome Lobby. (Only one workspace can be saved with a spatial anchor at a time.)
Load
Load molecules or structures into your workspace. Supported file formats include:
Structural Coordinate File Formats
.pdb, .cif, .mmcif, .pse, .nanome, .sdf, .mol, .xyz, .mol2
Electron Density Map File Formats
.ccp4, .dsn6
Trajectory/Molecular Dynamics File Formats
.dcd, .gro, .pqr, .trr, .xtc, .psf
Image File Formats
.png, .jpg, .jpeg, .pdf
Featured
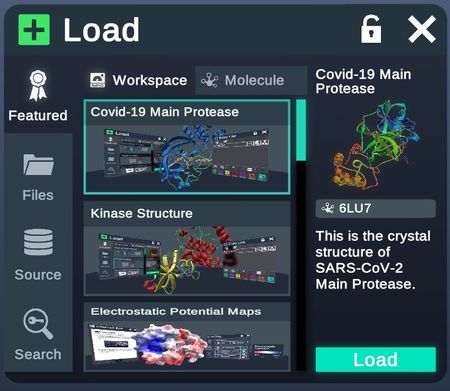
Workspaces that contain featured structures and tutorials.
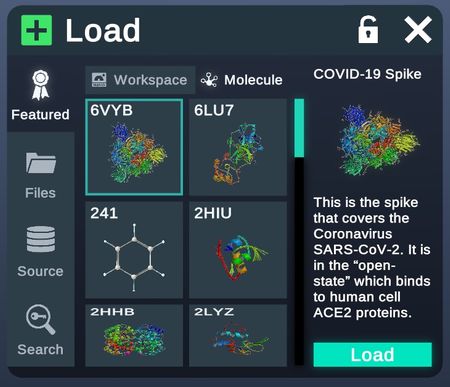
Molecules featured by Nanome for their interesting properties or background.
Database
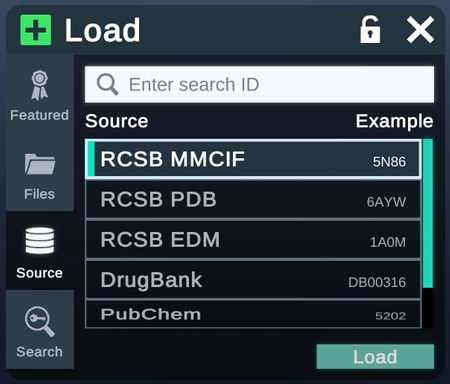
From the Database tab, structures can be directly loaded from RCSB, DrugBank (Academic and Enterprise only), or Pubchem (Academic and Enterprise only). Custom database can also be added by changing the config file.
Keyword
From the Keyword tab, structure can be loaded from a keyword search.
My Files
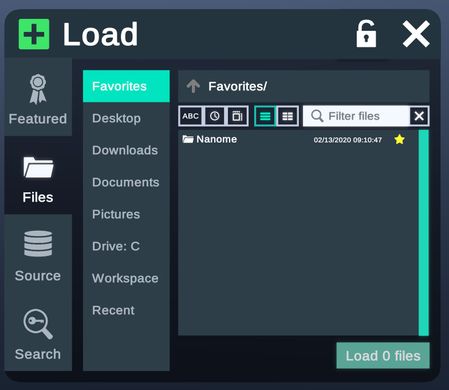
My Files tab allows users to load structures from their local directory.
Recent
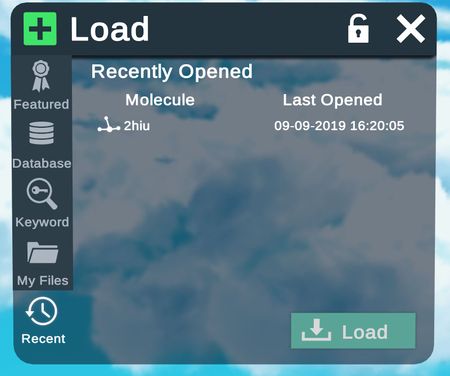
Recent files loaded in Nanome.
Cancel a load
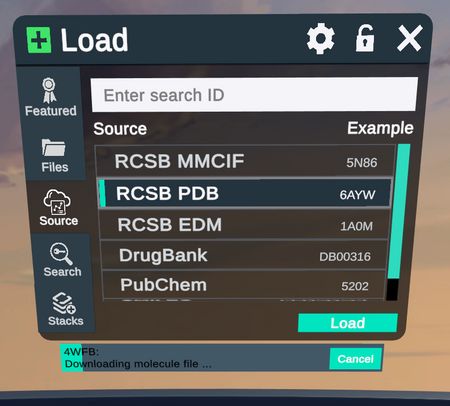 | 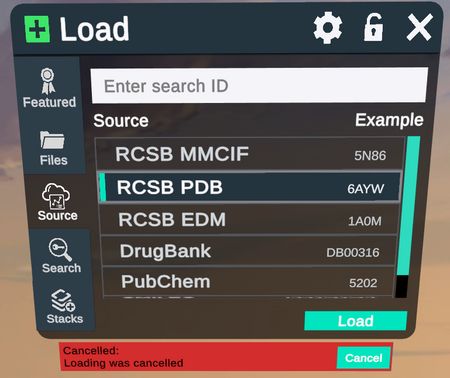 |
Cancel the loading of structures from the menu source tab, perfect for if your structure is taking too long to load or you made a mistake in the PDB code.
Stacks
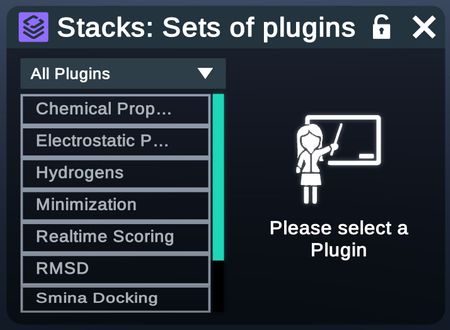
Please view the Nanome Stacks page to learn more.
Display
Atom
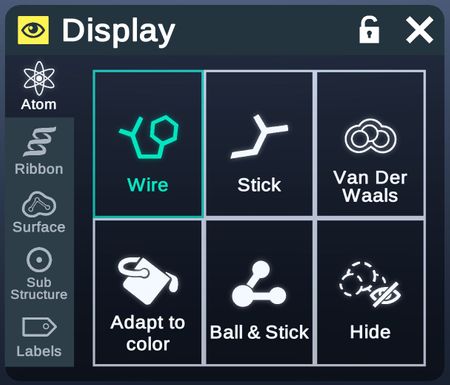
This tab contains options for the visual of atoms and bonds.
| Option | Description | Fig. |
|---|---|---|
 | Hide atoms and show bonds in thin wire visual. |  |
 | Show small atoms and bonds in volumetric stick visual. |  |
 | Show atoms in thin Van Der Waals visual and hide bonds. |  |
 | Show atoms with radius proportional to their b-factor value and hide bonds. | 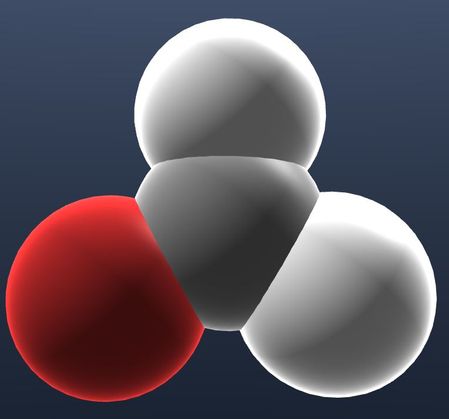 |
 | Hide all atoms and bonds. | 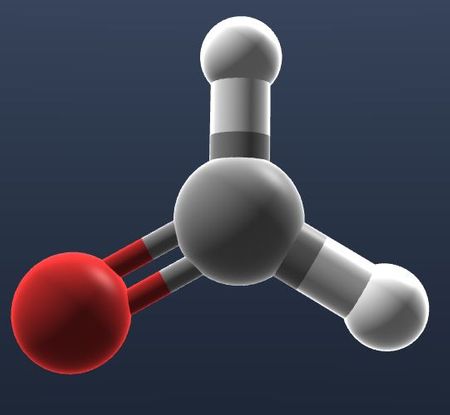 |
Ribbon
This tab contains options for the visual of ribbons.
| Visual | Description | Fig. |
|---|---|---|
 | Show ribbon in cartoon visual that highlight secondary structures using helix and arrows. | 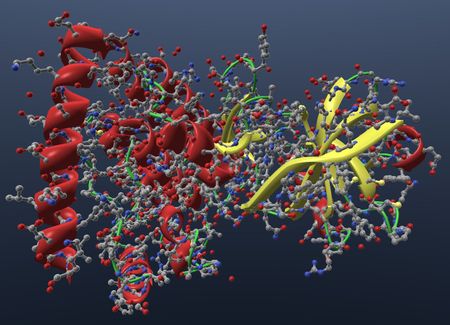 |
 | Show ribbon in uniform tube visual. | 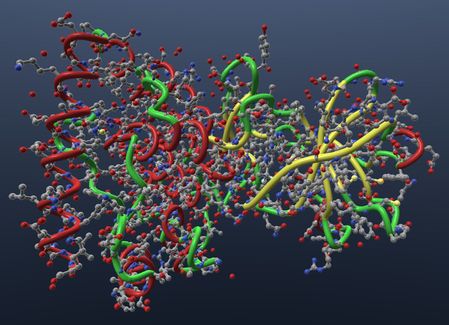 |
 | Show ribbon in tube with widths proportional to the atoms' b-factor value. |  |
Surface
Toggle the visibility of solvent-excluded-surface, change its opacity, or change the probe length (in Angstroms).
Mesh Surfaces (PCVR only)
If you are using PCVR, you also have the option to display a mesh surface. This option can be found in the Action Menu. Navigate to Display/Styles > Surface Styles, then check the "Mesh Surface" option.
Sub Structure
| Option | Description | Fig. |
|---|---|---|
 | Show hetatoms. | 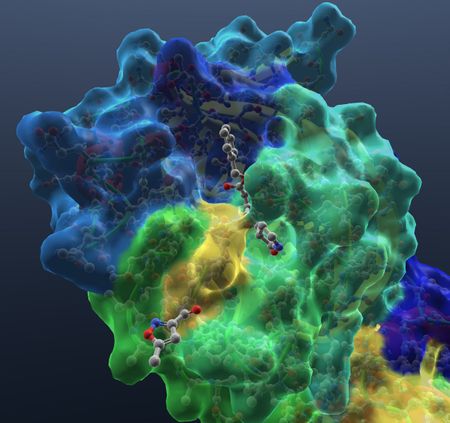 |
 | Show hetatoms, including water. | 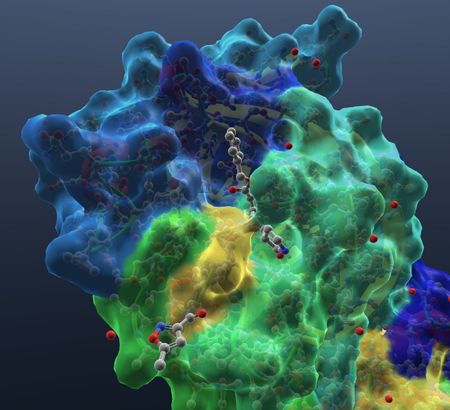 |
 | Show hydrogens. | 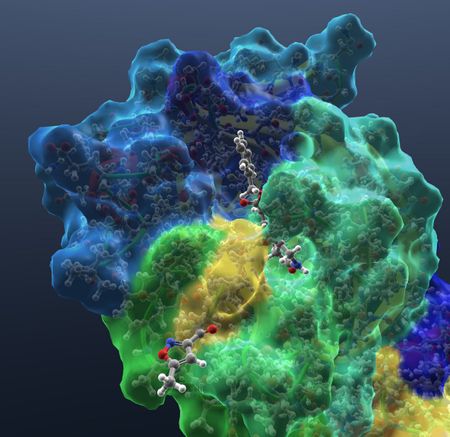 |
Labels
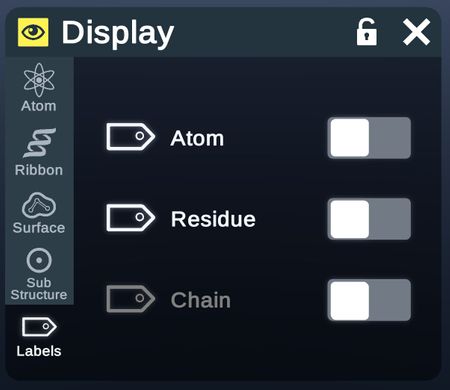
| Option | Description | Fig. |
|---|---|---|
 | Show labels on atoms. | 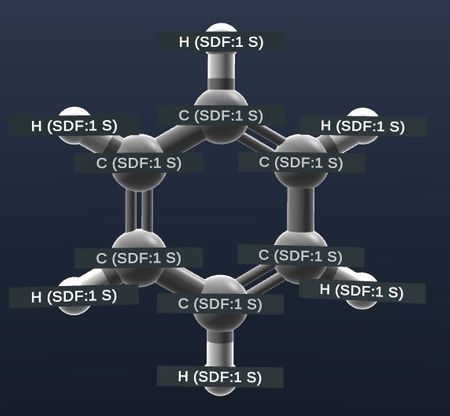 |
 | Show labels on residues. | 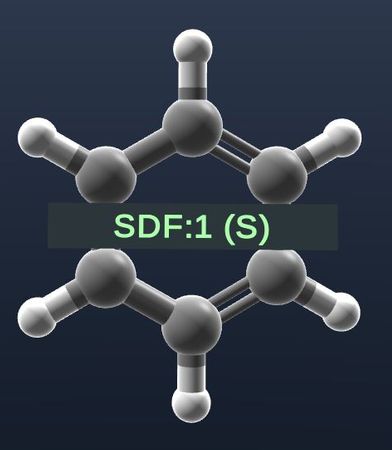 |
Color
Apply a solid color or a color scheme on the selected structures.
Picker
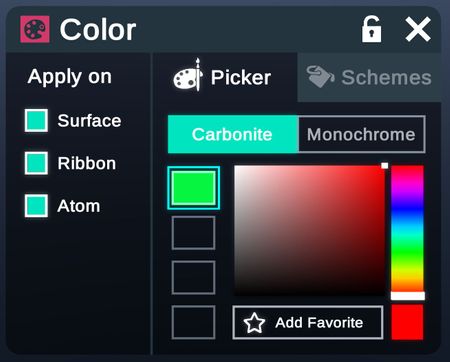
Colors picked in picker are applied in a solid style.
| Option | Description | Fig. |
|---|---|---|
 | Carbonite color applies only to carbons. | 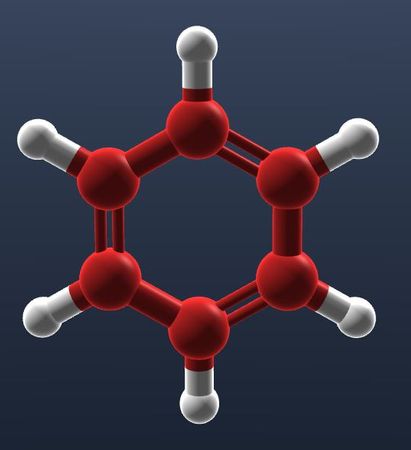 |
 | Monochrome color applies to all atoms indifferently. | 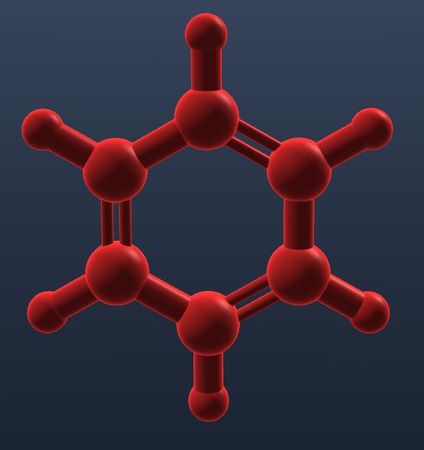 |
Schemes
Colors picked in schemes are applied in a certain pattern.
- YRB Hydrophobe
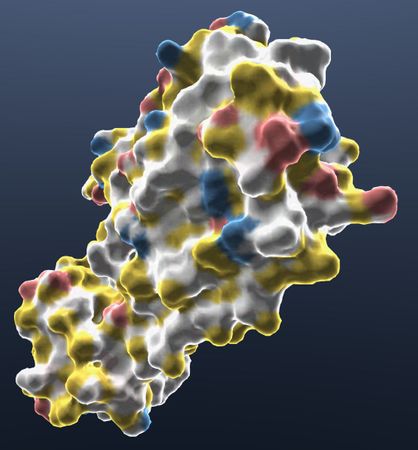
Color selected structures by their hydrophobicity with yellow, red, blue colors.
- IMGT
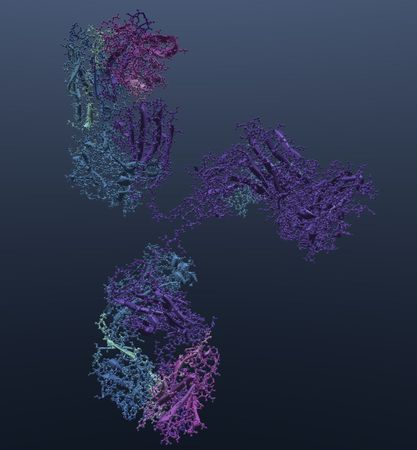
Color selected antibody structures by their regions.
Other available color schemes include coloring by occupancy, b-factor, rainbow and so on.
Modify
Edit
Edit the selected structures by applying actions from the buttons.
Compute
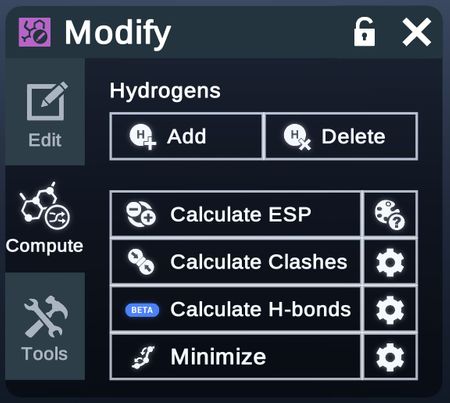
Apply certain computations on the selected structures and modify them according to the result.
- Calculate ESP
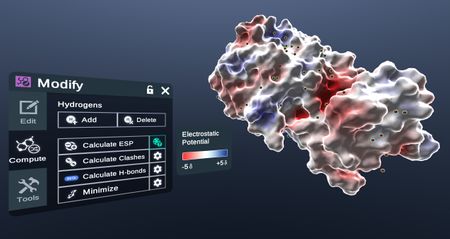
Generate the Electron Potential Map of the selected structure.
- Calculate Clashes
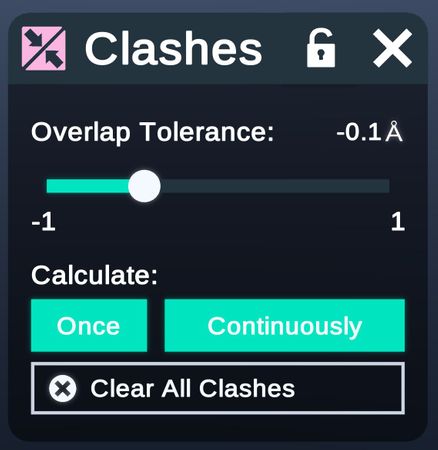
Config the tolerance of atom clashes.
- Calculate Hbond
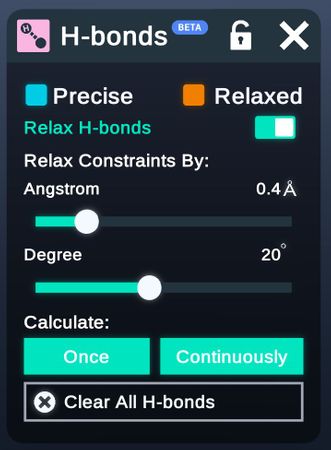
Config the threshold for generating h-bonds.
- Minimize
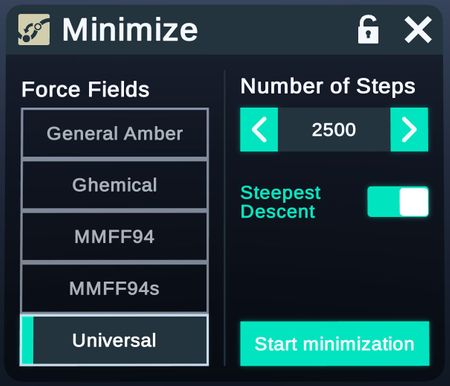
Config forcefield and step for minimizing potential energy of selected structures.
Tools
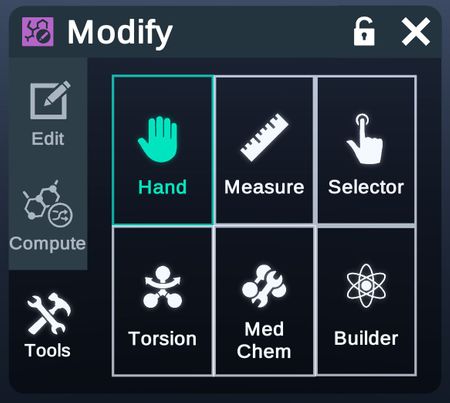
Switch to a certain tool to directly apply modification to the structures.
Mutate
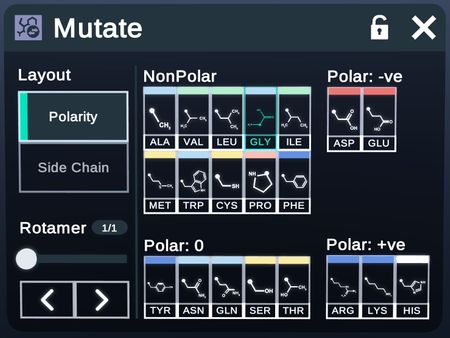
Mutate the R-groups of the selected amino acids.
Select
Quickly modify selection range using certain criteria.
Refine
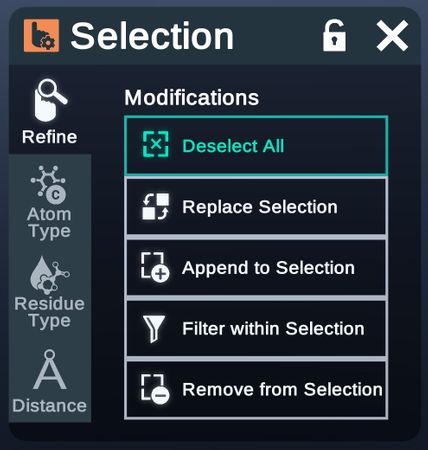
Refine selection by applying certain changes.
- Replace Selection
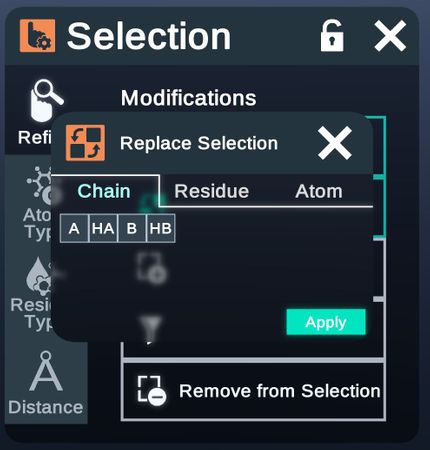
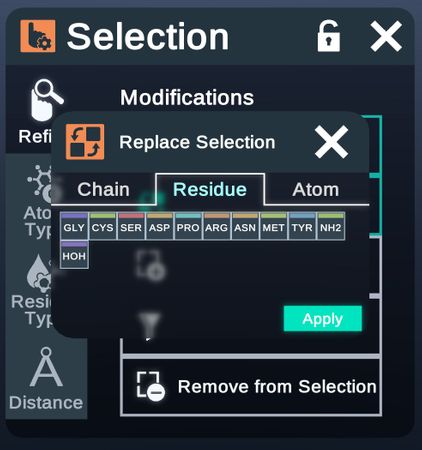
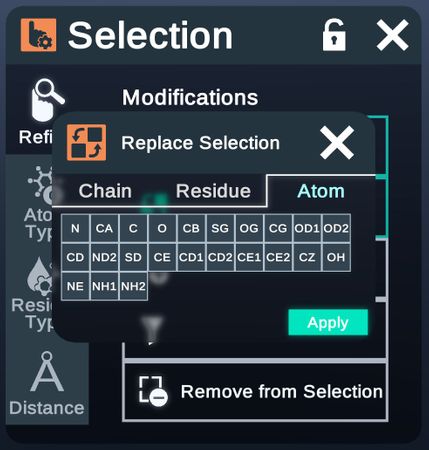
Atom Type
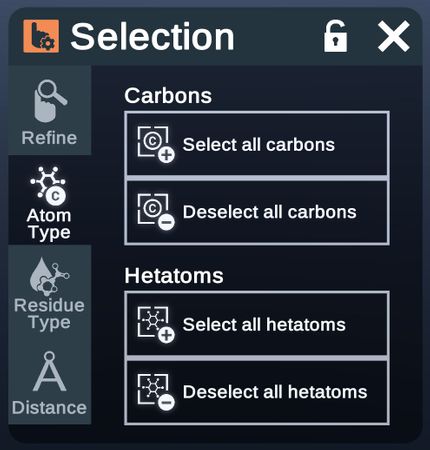
Select or deselect certain types of atoms.
Residue Type
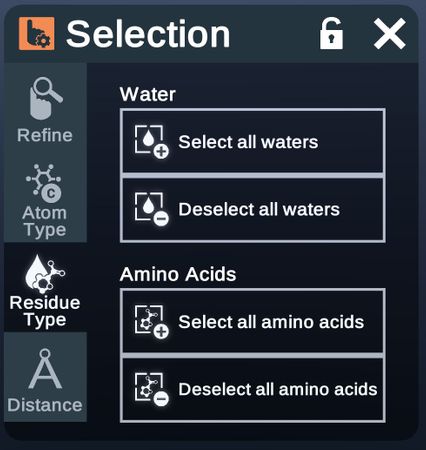
Select or deselect certain types of residues.
Distance
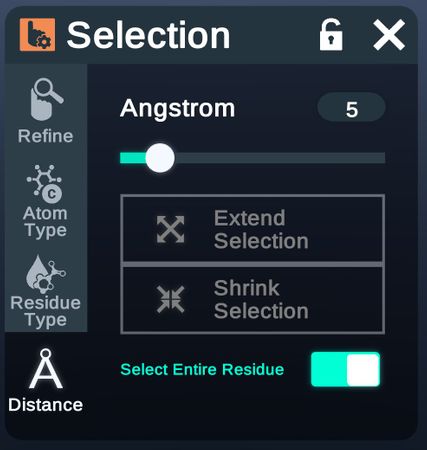
Expand or shrink the selection range by physical proximity.
Macro
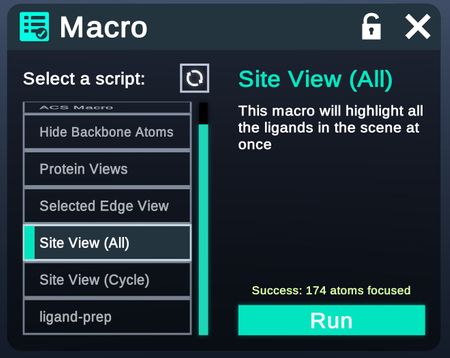
Please view the macros page to learn more.
Content Discovery
Tutorials of advanced concepts and features in Nanome, presented using spatial recordings.
Advanced Tutorials
See Content Discovery.
Spatial Recordings
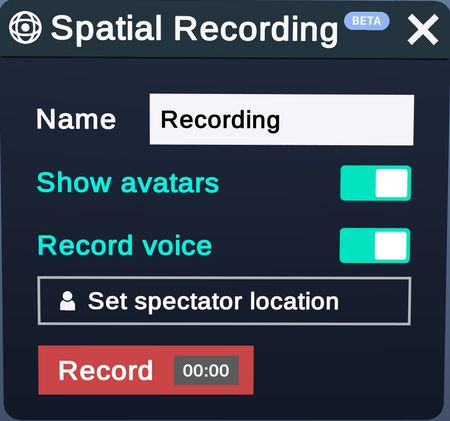
Record your entire workspace, including structures, menus, and virtual movements; then, playback at a later time. During the playback you can pause and even interact with the structures in the recording.
Spatial recordings can be accessed from either the Wrist Menu (see below) or the Action Menu.
As of April 2021 (Nanome 1.20), this feature is in BETA - in particular, spatial recordings created in version 1.20 will not be playable in future versions.