Appearance
Conformer Generator
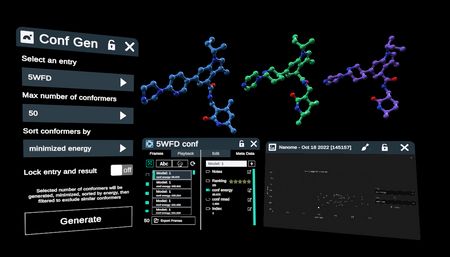
The Conformer Generator Plugin uses RDKit and UFF minimization to deliver a collection of diverse, low energy conformers of a selected “seed” ligand. 3D results are captured as frames in a single new entry in the workspace. The associated minimized energy and similarity to the seed ligand (RMSD) for the seed and all conformers is accessible in both the Advanced Menu - Frames and via the Data Table Plugin.
Potential uses
- Assess whether a protein bound ligand conformation is one of its low energy unbound conformations or if instead it is adopting a higher energy conformation
- Compare conformer energy and RMSD to that of the “seed” ligand
- Obtain low energy conformers for further analysis in docking protocols
- Visualize whether a ligand has many or few diverse low energy conformations available to it, to inform design strategy for more constrained or more flexible analogs
- Apply conformational analysis to selections for fragment screening and virtual screening as one parameter of hit quality
Instructions
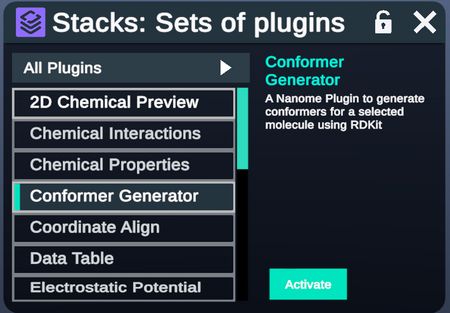
Access the Conformer Generator plugin from the Stacks Menu.
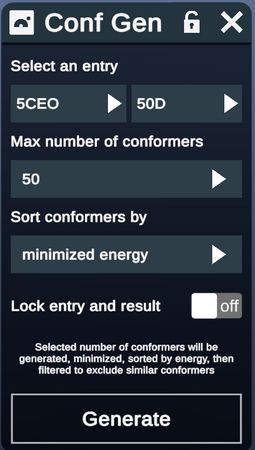
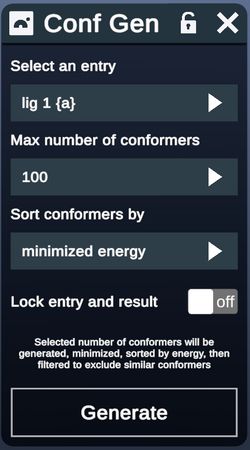
- In the upper left, the user can select either a protein entry containing a ligand or an individual ligand entry
- Next the number of conformers to be returned (50, 100 or 200 conformers) is chosen from the pulldown
- Users may optionally choose to have the conformer set locked with the seed structure
- Clicking Generate runs the plugin. During calculation, this button reads “Generating”
- Once complete, the “Generating” button returns to “Generate” state and a new entry appears in the entry list
- The new entry of conformer frames has associated meta data of conformation energy and RMSD to seed ligand
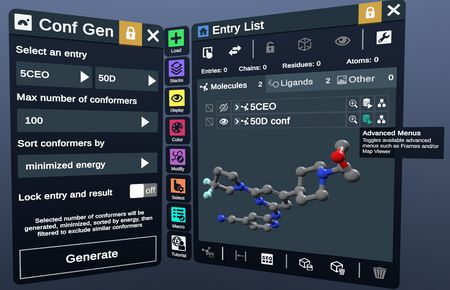
The conformer entry of frames opens in the workspace, and the new entry list item appears, with a link to the Advanced Menu for navigating frames.
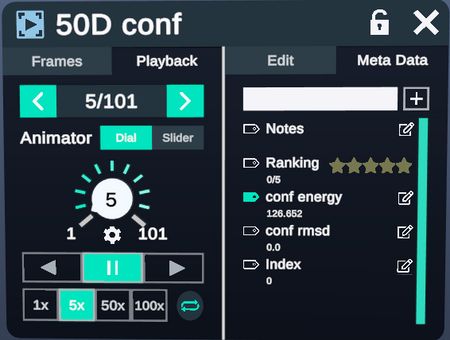
Advanced Menu allows sorting and playing back through the conformers, as well as viewing the energy and RMSD of each conformer.
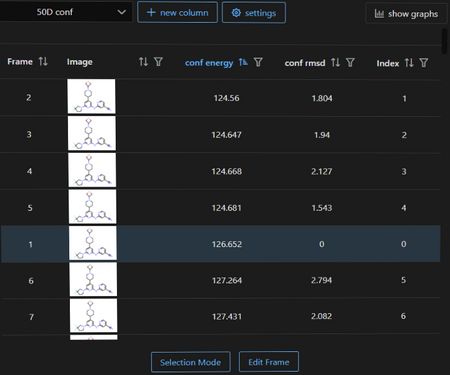
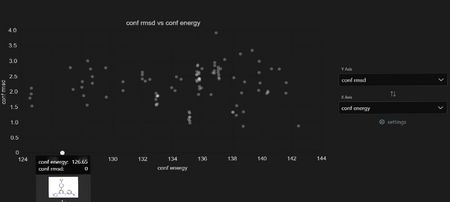
The Data Table plugin can also be used to view results and is accessible through the Stacks Menu. Energy and RMSD to seed ligand may be sorted and filtered in the table, and plotted graphically. Note the seed ligand at RMSD=0 highlighted here.